Real time Polymerase chain reaction (RT PCR)
Nano to Micro/Macro (Biomolecules and biomaterials analysis)
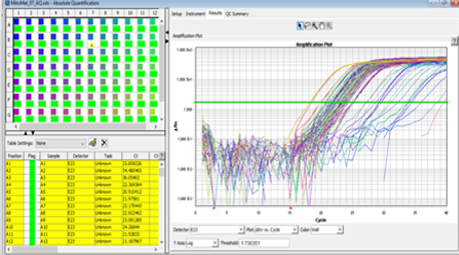
Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected throughout the PCR process, rather than at the end of the PCR. In real-time PCR reactions are characterized by the point in time during cycling when amplification of a target is first detected rather than the amount of target accumulated after a fixed number of cycles.
The main chemistries used to detect PCR products are:
PCR primer- and probe-based detection chemistries (e.g. TaqMan Chemistry, molecular beacons, scorpion PCR primers): these chemistries use some form of fluorescence quenching to ensure that target-specific fluorescence is detected only when amplicon from the product of interest is present. The PCR primer or target-specific oligonucleotide probe is labeled with a reporter fluorophore and its fluorescence is quenched when the specific target DNA sequence is not present: this quenching is accomplished by covalently attaching a quencher molecule to the DNA primer or probe in combination with some mechanism by which the reporter and quencher are separated when the primer or probe binds to its specific target sequence. SYBR® Green Dye Chemistry: SYBR® green dye molecule binds non-specifically to double-stranded DNA (dsDNA). SYBR® Green exhibits little fluorescence when it is free in solution, but its fluorescence increases up to 1,000-fold when it binds to dsDNA. As the PCR progresses, more amplicons are created. Since the SYBR Green dye binds to all double-stranded DNA, the result is an increase in fluorescence intensity proportionate to the amount of PCR product produced.